
Synthetic Biology: Life, Remixed
Like engineering, synthetic biology involves building new biological systems from units and modules. Scientists have already produced organisms that make drugs or biofuels, and have pieced together a synthetic organism using only genetic data. In collaboration with other fields, synthetic biology could transform biotechnology and generate useful insights into the foundations of life.
The goal of synthetic biology is to produce novel living systems with desirable properties.
Biotechnologists have long been able to take an organism and modify it to make it more useful to humans. By inserting genes here or cutting genes there, it has been possible to turn microbes into tiny protein factories, design better crop varieties and study human disease in animal models. Now, scientists working in the field of synthetic biology are exploring ways in which technology can not only alter life, but create it.
The fundamental concept of synthetic biology is not unlike how an engineer might assemble a chemical plant — but instead of pumps, reactors and filters, synthetic biologists work with a toolbox full of genes, proteins and other biomolecules1.
Their goal is to produce new living systems that, when deployed in the lab under controlled conditions, have desirable properties — for example, the ability to make complex products and materials or detect environmental changes. Although the field of synthetic biology is still young, its proponents expect it to lead us to an exciting new era in biotechnology.
Cell upgrade
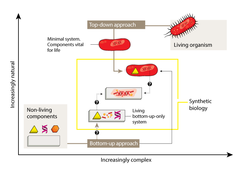
The most fruitful line of research has involved a ‘top-down’ approach — starting with an unmodified or simplified cell and adding foreign elements or modules (Fig. 1). These might consist of genes encoding proteins that synthesize a molecule of interest in a sort of microscopic assembly line, or that cause a detectable change in response to an incoming signal. Although top-down synthetic biology is more complex than biotechnology strategies that modify only single genes, the tools and techniques are generally the same.
Even so, building self-contained modules with designer properties, and then integrating them into host cells without adverse effects, is an enormous challenge. Top-down synthetic biology already has several success stories, however. For example, researchers introduced genes into yeast cells to cause them to produce artemisinic acid2, the precursor of a key antimalarial drug that is in short supply. This system is now being developed with the aim of mass-producing cheap anti-malarial drugs. Another example is the introduction of networks of genes that enable yeast, bacteria or algae cells to produce biofuels, although these pilot studies have also not yet reached the level of efficiency to move to large scale production3.
One remarkable achievement in top-down synthetic biology has been the production of viable, synthetic bacteria using only genome data as a blueprint. With blocks of DNA, researchers assembled the genome of Mycobacterium genitalium and transferred it to another species of bacterium that had been emptied of its own DNA. They found that the recipient yielded healthy synthetic offspring with the characteristics of the genetic donor4.
This work is an impressive technical achievement in itself. More importantly, as only around 80% of genes in the donor species are essential5, it is also an important step on the path to constructing a simplified cell. Over billions of years of evolution, even the simplest cells have achieved a degree of complexity that is beyond the immediate needs of synthetic biology, and is likely to confound the attempts of scientists to make much progress. By sidestepping this complexity, minimal systems have the potential to provide a series of well-understood starting points for future efforts.
Back to basics
The problems posed by the complexity of natural organisms can also be avoided through a ‘bottom-up’ approach. Instead of starting with existing biological systems, this approach involves creating entirely new modules and systems based on fundamental biological principles (Fig. 1), drawing on a combination of natural, modified and synthetic biomolecules as building blocks. The creation of bottom-up systems is inherently more challenging than the top-down strategy — indeed, it remains unclear whether will ever be possible to create life using only a bottom-up approach.
In the meantime, some scientists are opting to work with extracts derived from ruptured cells, which contain all the necessary machinery of life but which can be manipulated without the need to preserve the integrity and survival of the host cell6. These ‘cell-free’ systems offer a promising testing ground for synthetic biology. However, the contents of such extracts are poorly defined, removing an important element of experimental control, and by nature they are not whole, self-replicating biological systems.
The development of ‘proto-cells’ that mimic the fundamental structural and functional principles of real cells is therefore a major goal of bottom-up synthetic biology. The contents of a cell are encased in an outer membrane, and usually further segregated according to their functions within specific structures, such as the nucleus and endoplasmic reticulum. Scientists are experimenting with different membrane formulations in an effort to imitate this segregation.
Early work in this area has shown that biological reactions such as DNA replication can occur in basic membrane-bound structures7, and that introducing protein pores to artificial membranes can allow the controlled exchange of molecules with the environment, as happens in nature8.
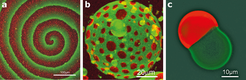
Yet for us to make much progress with the bottom-up approach, minimized systems will need to be able to adapt to changes in their immediate environment, by establishing organizational patterns and maintaining equilibrium. Researchers have identified several striking examples of molecular self-organization9 (Fig. 2), but these are far more primitive than those seen in even the simplest life forms. So far, only a few natural phenomena have been mimicked using bottom-up approaches.
Bringing the concept to life
One of the great challenges for both top-down and bottom-up synthetic biology is identifying the minimal set of components needed for core processes, such as cell division and the generation of energy from nutrients, to take place. Uncovering these primal elements of cellular function, and then learning how to fit them together into new scaffolds, will involve both top-down and bottom-up approaches, and will take years of hard work by scientists in a broad range of disciplines. However, the ultimate rewards for this effort may be a richer understanding of what it means to be alive.
By reproducing simple biological phenomena in vitro, researchers at the Max Planck Institute of Biochemistry have made important progress in producing biomimetic bottom-up systems. Vogel et al. (eLife 2013;2:e00116) have reconstituted a minimized actin cortex, the mesh that supports cell membranes. Meanwhile, Zieske and Schwille (Angew. Chem. Int. Ed. 52 459-462; 2013) have shown that proteins required for cell division in bacteria can self-organize to form dynamic patterns and oscillations in artificial membrane compartments.
References
1Rollié S et al., Chem. Eng. Sci. 69, 1 (2012)
2Ro D-K et al., Nature 440, 940 (2006).
3Peralta-Yahya PP et al., Nature 488, 320 (2012).
4Gibson DG et al., Science 329, 52 (2010).
5Glass JI et al., Proc. Natl. Acad. Sci. USA 103, 425 (2010).
6Hodgman CE & Jewett MC, Metab. Eng. 14, 261 (2012).
7Kurihara K et al., Nat. Chem. 3, 775 (2011).
8Noireaux V & Libchaber A, Proc. Natl. Acad. Sci. USA 101, 17669 (2004)
9Loose M et al., Science 320, 789 (2008).
Content taken from Max Planck Society.

