Theory of Self-organizing Synthetic Biosystems
Work Package S1
An essential step for the development of a structured model library lies in the identification and characterization of functional modules necessary for the construction of self-reproducing proliferating systems. This is done in two ways embodied by the tasks of Work Package S1: The first way is to analyze unstructured or weakly structured theoretical whole-cell models taken from literature, to determine the functional modules hidden in these theoretical models as well as their essential properties and replace them by experimentally validated biochemical systems possessing these properties. The second possibility is to start with subsystems of artificial cells that have been studied in experiments in an isolated way, i.e. decoupled from the other parts of a cell. The subsystem models can be aggregated to models of cell-like entities by defining interfaces for information exchange between the subsystems. The aggregated system model allows to assess if the constructed artificial cell has the desired properties, or if additional functional modules are required, e.g. for coordination and synchronization of the subsystems.
The final goal of Work Package S1 is the implementation of a validated library of whole-cell models as well as of cell parts or functional modules in a simulation program. The library will serve as an in-silico bottom-up cell designer to support the development of biological systems with defined functionalities.
An essential step for the development of a structured model library lies in the identification and characterization of functional modules necessary for the construction of self-reproducing proliferating systems. In Work Package S1, this is done in two ways: The first way is to analyze unstructured or weakly structured theoretical whole-cell models taken from literature, to determine the functional modules hidden in these theoretical models as well as their essential properties, and to look for experimentally validated biochemical systems possessing these properties. The second possibility is to start with subsystems of artificial cells that have been studied in experiments in an isolated way, i.e. decoupled from the other parts of a cell. Models for these subsystems have to be developed within Work Package S1 in cooperation with the MaxSynBio partners, using their expertise and experimental data. The subsystem models can then be aggregated to models of cell-like entities by defining interfaces for information exchange between the subsystems. The figure shows an example of such an aggregated system, whose desired functionality is to grow and divide. The aggregated system model allows to assess if the constructed artificial cell actually has the desired properties, or if additional functional modules are required, e.g. for coordination and synchronization of the subsystems.
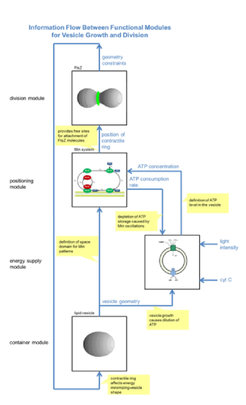
Work Package S1 adds a system-wide perspective to the MaxSynBio project. While the other work packages focus on a detailed understanding of partial aspects of artificial cells like membrane formation, motility, signaling, or division, Work Package S1 intends to form a system theoretic umbrella over these activities. By combining the results of the partners to whole-cell models, it shifts the focus from the internal properties of single functional modules to the interaction between different functional modules. This allows comparing alternative approaches for the same functionality in the context of a complete artificial cell, e.g. different mechanisms for positioning a contractile ring.
The combination of various functional modules to an artificial cell may be a tremendous task in experiments, but is much easier in the simulations done within Work Package S1. Therefore, Work Package S1 also has an extrapolating character, as it attempts to make predictions on artificial biological systems that during the runtime of MaxSynBio cannot be implemented in experiments due to time limitations or other constrictions.
